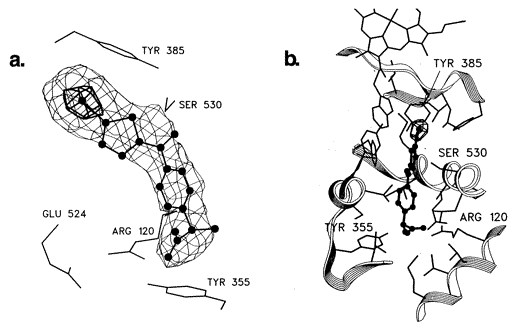
Binding of iodosuprofen to PGHS-1

Fig.9 Stereoviews of the difference electron density corresponding to the iodosuprofen molecule bound to PGHS. The carboxylate group of the inhibitor engages in a salt bridge with Arg-120 and lies within hydrogen-bonding distance of the hydroxyl of Tyr-355. Glu-524 is also engaged in a salt bridge interaction with Arg-120. The iodothiophene moiety packs directly under Tyr-385. In this view, Ser-530 lies behind the inhibitor; its side chain hydroxyl is positioned to make a hydrogen bond with the ketone oxygen of the inhibitor. (b) Stereoview of the bound inhibitor showing the protein backbone and side chains making up the active site cavity. A portion of the heme group is seen at the top of the figure (8)
Binding of iodosuprofen and iodoindomethacin to the cyclooxygenase channel
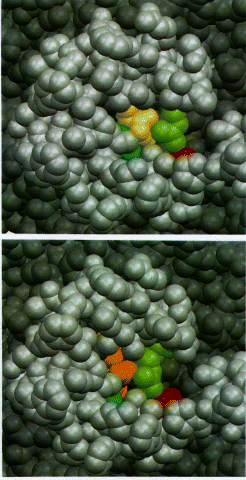
Fig.10 Space-filling models of the mouth of the cyclooxygenase channel viewed from the membrane. In top and bottom, the catalytic domain is shown in dark gray while the amphipathic membrane binding domain is shown in light gray. Arg-120 (green) and Tyr-355 (blue) form part of the opening; Glu-524 is shown in red. (Top) Iodosuprofen (yellow) is shown bound within the channel. in (bottom) iodoindomethacin (orange) is shown bound. The membrane binding domain and the residues Arg-120 and Tyr-355 block the access of the bound drugs to the exterior of the enzyme toward the membrane side. Ligand binding and dissociation should involve structural alterations of the mouth of the cyclooxygenase channel. (8)