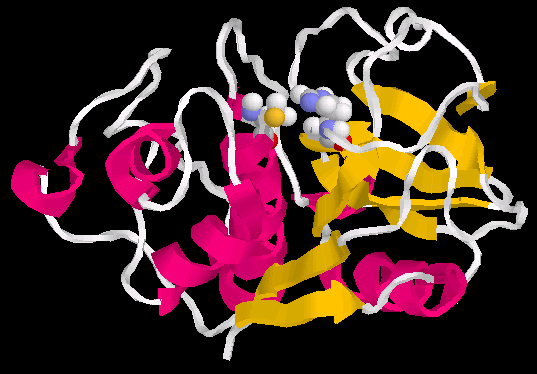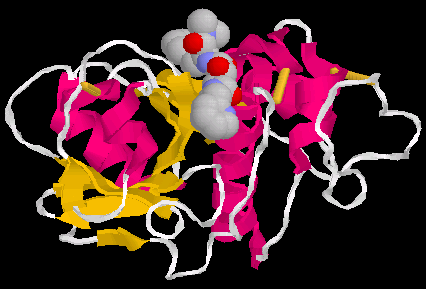MOLECULAR MODELLING AND STRUCTURAL ANALYSIS OF PAPAIN

|
||||||||||||||
|
INTRODUCTION:
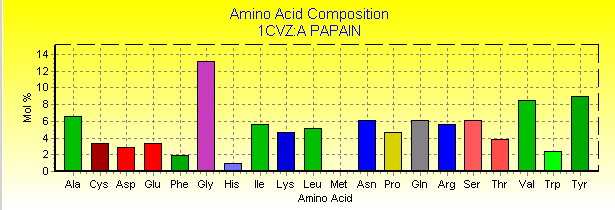
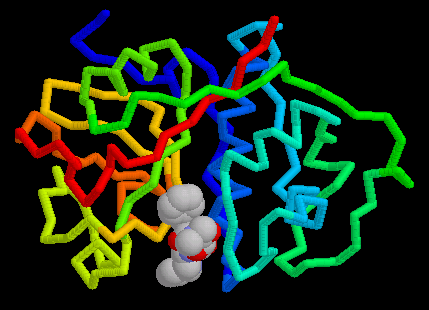
Papain is an endolytic cysteine protease which is isolated from papaya latex. It belongs to the peptidase family - C1, and is noted for its wide specificity. It preferentially cleaves peptide bonds involving basic amino acids, particularly arginine, lysine and residues following phenylalanine(1). A three-dimensional structure has been indicated by Wolthers et al (1970). The molecule consists of a single, folded polypeptide chain of 212 amino acid residues(Fig. 1a,b below), containing 3 disulphide bonds(Fig.8) and one free functional -SH group at the active site( Smith et al. 1975). Many other plants contain cysteine endopeptidases, all of which seem to be homologs of papain, often with similar specificity. Examples are calotropin from the madar plant (Calotropis gigantea), phytolacin from the pokeweed (Phytolacca americana) and mexicanain from Pileus mexicanus (2). The uses for papain are diverse and thus it makes it an extremely valuable enzyme(3). Table 1, below provides classification and structural summary of papain(pdb1cvz)(4)
Papain is well known for it's hydrolase activity at the hinge site of immunoglobin. This is an interesting activity considering it is found in the juice and leaves of an unripe papaya (Carcia papaya). This enzyme is believed to be produced as a defense against having the fruit eaten before its seeds have matured(5).
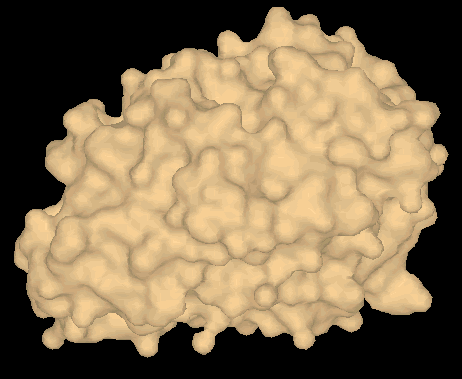
Fig2 shows a 3D model (surface view of papain(pdb 1cvz)(5).
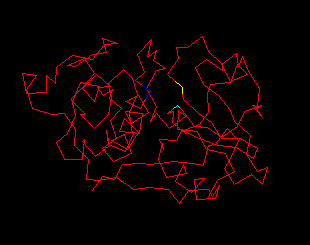
Fig. 3 below shows the 212 residues
of papain(1cvz) .(4)
The PDB structure of papain was determined
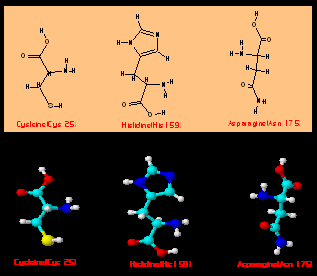
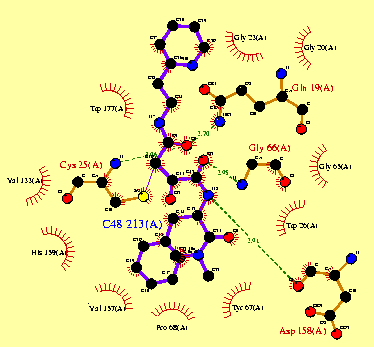
by x-ray diffraction(ray crystallography) (7). Figure 4a shows the secondary structure of papain(pdb1cvz)
and the amino acids(Cys 25, His 159 and Asn 175) which are part of the catalytic
center. Fig. 3b shows both 2D and 3D structure of these amino
acids.
Figure 5a and 5b below, shows the secondary structure of papain (1cvz) , in which the alpha helices, and beta sheets are colored red and yellow respectively(5). |
||||||||||||||
|
formatic enough,
>maybe they should start new threads.
>
>Here's one:
>
>If we're going to talk about "what bioinformatics needs" (quoting
>Jon), it seems that we should attempt to nail down what
>"bioinformatics" _is_.
>
>I'll offer a starting point[1]: "bioinformatics" is really just
>computational biology under a newer, more media-friendly name -- that
>is, it's a largely content-free buzz-phrase invoked to describe some
>combination of something to do with computers, and something to do
>with biology.
>
>Dissenting opinions welcome. 8^)=
>
>john.
>
Papain is an
References
http://www.d-trends.com/webs/ics_preface.html
>maybe they should start new threads.
>
>Here's one:
>
>If we're going to talk about "what bioinformatics needs" (quoting
>Jon), it seems that we should attempt to nail down what
>"bioinformatics" _is_.
>
>I'll offer a starting point[1]: "bioinformatics" is really just
>computational biology under a newer, more media-friendly name -- that
>is, it's a largely content-free buzz-phrase invoked to describe some
>combination of something to do with computers, and something to do
>with biology.
>
>Dissenting opinions welcome. 8^)=
>
>john.
>
Papain is an
References
http://www.d-trends.com/webs/ics_preface.html