HYDROPHOBICITY OF PAPAIN(1CVZ)
The hydrophobic -hydrophilic
interaction of amino acids in the side chain seems to be the major thermodynamic
force(s), that drives protein folding.(1).
In the context of protein structures, several amino acid side chains are,
to varying degreees, hydrophobic. The most hydrophobic of the amino acid side
chains are those of alanine, valine, leucine and isoleucine(2). Fig
1, and 2 ,below shows hydrophobicity of papain(1cvz) (3).
|
|
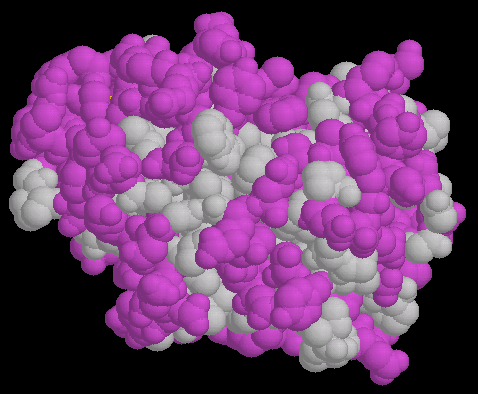
FIG.1
Fig. 1 show a space fill model of papain(1cvz) in which the hydrophobic
amino ancid are colored purple. The backbone oxygen and nitrogen of the
residues with the hydrophobic sidechain are colored gray like the side chain.They
are of course polar thought most of their hydrogen bonding requirements are
typically satisfied within the backbone.
|
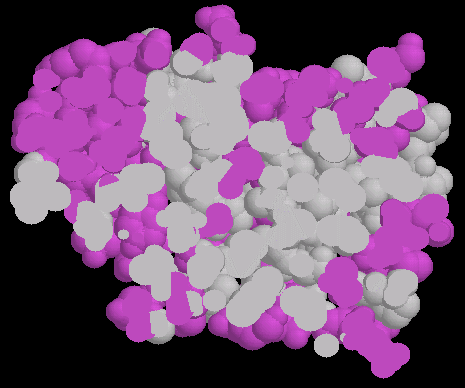
FIG.2
Fig. 2 shows a half-slice of the papain(1cvz)
molecule shown in fig. 1.
This was achieved by using the slab mode of Chime software and is very useful
to see how the hydrophobic residues are distributed. |
HYDROPHATIC PROFILE OF PAPAIN(1CVZ)
Hydropathy plots allow for
the visualization of hydrophobicity over the length of a peptide sequence.
A hydropathy scale which is based on the hydrophobic and hydrophilic properties
of the 20 amino acids is used. As show in Fig. 3 below a moving "window"(white
line parallel to y axis) determines the summed hydropathy at each point in
the sequence (Y coordinate). These sums are then plotted against their respective
positions (X coordinate). Such plots are useful in determining the hydrophobic
interior portions of globular proteins as well as determining membrane spanning
regions of membrane bound proteins(3.)
Fig. 3 below shows a
graphical representation of the hydrophatic properties of papain(1cvz). The
moving window is align with v, which
is part of the first helix of the papain molecule. The sequence of the first
helix in papain(1cvz) is underline in the Fasta format below and corresponds
to the amino acid valine.
IPEYVDWRQKGAVTPVKNQGSCGSCWAFSAVVTIEGIIKIRTGNLNQYSEQELLDCDRRSYCG
CNGGYPWSALQLVAQYGIHYRNTYPYEGVQRYCRSREKGPYAAKTDGVRQVQPYNEGALLY
SIANQPVSVVLQAAGKDFQLYRGGIFVGPCGNKVDHAVAAVGYGPNYILIKNSWGTGWGEN
GYIRIKRGTGNSYGVCGLYTSSFYPVKN
|
Fasta Format of
Papain(1cvz) shown above
The hydrophobicty character
is the property of a side chain to be less soluble in water than in a nonpolar
solvent. For example, the energy to transfer the side chain of each amino
acid from a polar to a non polar solvent (water to ethanol) has been measured.
An hydrophobicity scale has been derived (3).
G -0.4
|
Q -3.5
|
S -0.8
|
Y -1.3
|
A 1.8
|
K -3.9
|
T -0.7
|
W -0.9
|
V 4.2
|
H -3.2
|
D -3.5
|
C 2.5
|
L 3.8
|
R -4.5
|
E -3.5
|
M 1.9
|
I 4.5
|
F 2.8
|
N -3.5
|
P -1.6
|
Hydrophobicity
scale
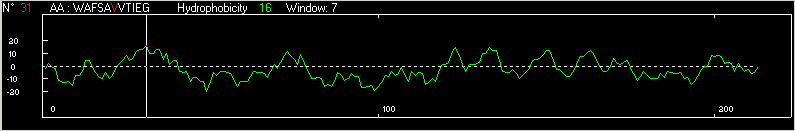
Fig. 3 Hydrophobicity of papain(1cvz)
HYDROPHOBICITY OF THE HELICES
It is often useful to examine
the relative hydrophobicity or hydrophilicity values of the amino acids in
a protein sequence. Since hydrophobic residues tend to be more buried in
the interior of the molecule and hydrophilic residues are more exposed to
solvent, a profile of these values can indicate the overall folding pattern.
Specifically, a long stretch of hydrophobic residues can indicate a buried
-strand, and a short spike of hydrophiliicity can indicate a turn. The hydrophobic interaction have the major influence in
protein confirmation.
|
SUMMARY OF THE HELICES IN PAPAIN
Helix
Number |
Start
|
End
|
Residues
per turn
|
Sequence |
1
|
25
|
42
|
3.67
|
CWAFSAVVTIEGIIKIRT
|
2
|
56
|
56
|
3.71
|
EQELLDC
|
3
|
62
|
64
|
-
|
GCN
|
4
|
68
|
77
|
3.61
|
PWSALQLVAQ
|
5
|
118
|
127
|
3.57
|
QGALLYSIAN
|
6
|
139
|
142
|
3.86
|
KDFQ
|
7
|
199
|
201
|
-
|
VCG
|
Table: 1
Table 1 shows the
sequences of the seven residues in papain(1cvz)
|
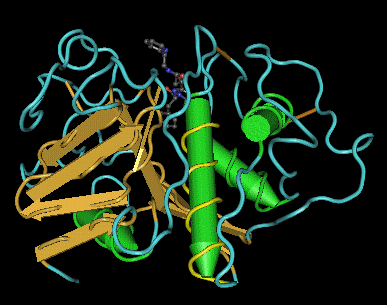
Fig. 4
Fig. 4 shows a 3D model of papain(1cvz) in which the first
helix has been rotated for easy visualization. Helices with less
than a turn is is
not visible.
|
|
|
|
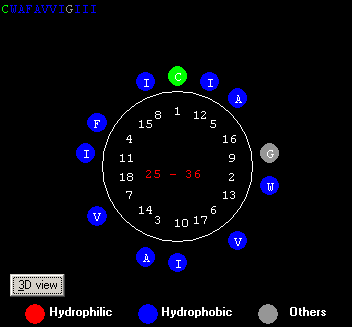
Fig.5
Fig. 5 above shows a Helical wheel of the first
helix in papain, colored by hydrophobicity. Hydrophilic= 33%;
Hydrophobic=55%;Others=12% .
|
Fig.6
Fig. 6 shows the hydophobic amino acids for the first
helix. This helix appears to be found embedded away from the surface because
of the significant hydrophoic residues.
|
Fig. 7 below (right) shows the Graphical
Contact of the residues(WA) in the first helix of papain sequence(CWAFSAVVTIEGIIKIRT) (1cvz). The legend on the
left indicates there are 9 hydrophobic interactions, and four hydrogen bonds,
two on the side chain and two on the main chain). By using the sliding window
the contact for each residue can be viewed.(11)
|
|
![]()

![]()
