Structure Neighbors and 3D alignments for 1CVZ (papain) Chain
A(Size=212)
A search
for protein with similar structure to the of 1CVZ chain A was carried
out. The result obtained ranged from 6.8% - 100% sequence similarity. 98.5
- 100% similarity was obtained with otherr cysteine proteinase obtained from
Carica Papaya. However,since the objective is to
check for sequence similarity in organisms other than Carica Papaya,only those
are shown in table 1 below.
ID
|
Z-score
|
RMSD(Å)
|
Seq.(%)
|
Aligned/size
|
Gap
|
Name
|
Source
|
1CVZ:A
|
QUERY |
-
|
-
|
-
|
-
|
Papain
|
Carica Papaya
(1)
|
1MEM:A
|
7.2
|
1.1
|
44.0
|
207/215
|
12
|
CathepsinK
|
Kiwi Fruit
or Chinese Gooseberry (2) |
2ACT
|
7.1
|
0.9
|
48.5
|
204/220
|
20
|
Actinidin
|
Homo sapiens (3)
|
1F2C:A
|
7.1
|
1.2
|
36.5
|
203/215
|
19
|
Cruzain
|
Trypanasoma cruzi (4) |
8PCH:A
|
7.1
|
1.4
|
42.6
|
202/233
|
21
|
Cathepsin
H |
Sus scrofa(pig)
(5)
|
Table: 1
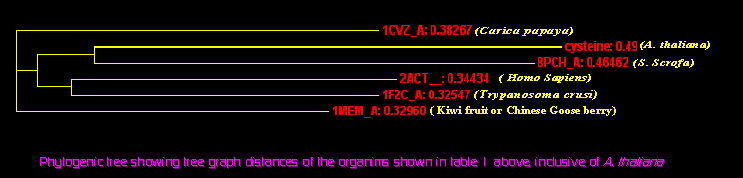
|
The tree was generated
in Clustalw tool available at EBI (7).
|
SEQUENCE
ALIGNMENT
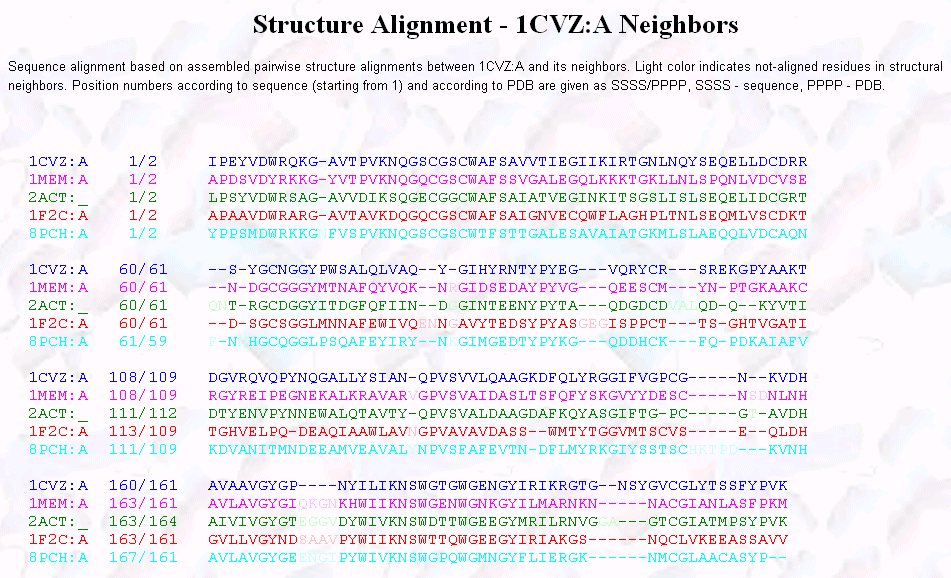
3D STRUCTURAL ALIGNMENT OF 1CVZ(PAPAIN) WITH CATHEPSIN
K (Fig.1 below)12
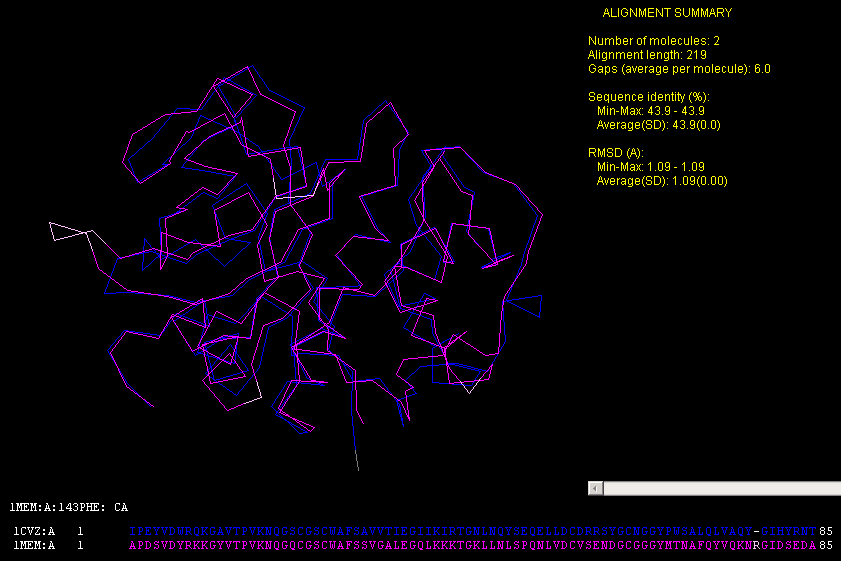
Fig. 1
3D STRUCTURAL ALIGNMENT OF 1CVZ(PAPAIN) WITH 2ACT(Fig.2
below)13
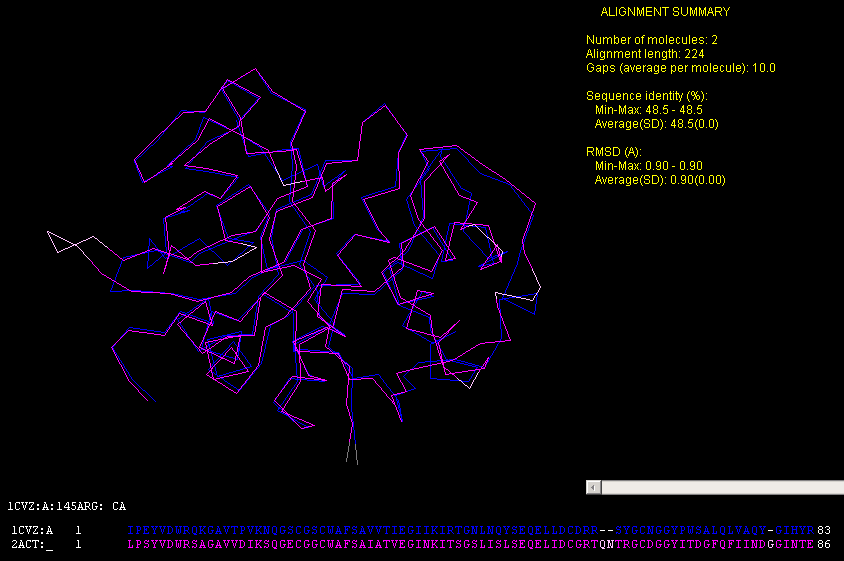
Fig. 2
3D STRUCTURAL ALIGNMENT OF ALL THE
PROTEINS SHOWN IN TABLE 1 ABOVE(Fig.3 below)
Fig.3
|
![]()

![]()
